Cross_Docking
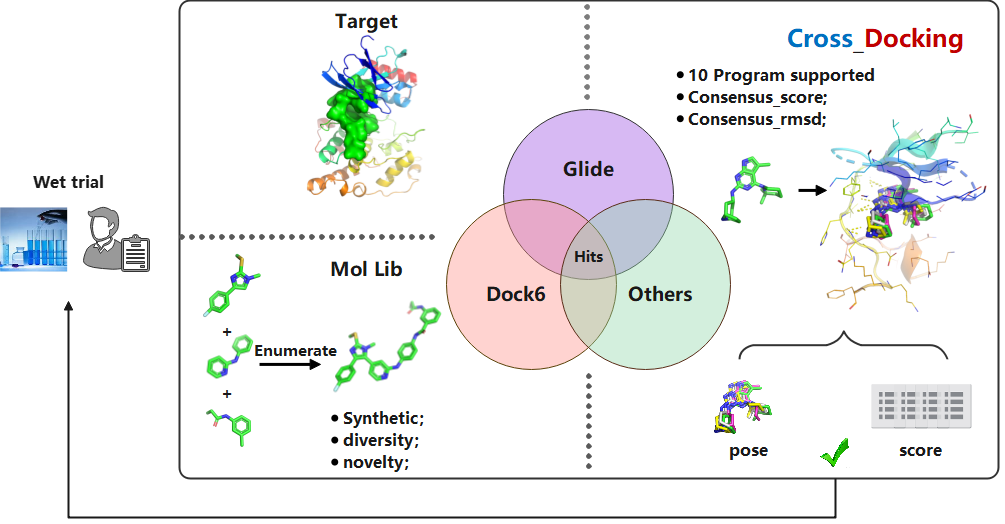
The CrossDock module addresses a fundamental challenge in molecular virtual screening: the accurate prediction of ligand binding poses. Inaccurate pose prediction is a critical source of error, often leading to misleading binding scores that degrade ranking performance and significantly increase the false positive rate of screening campaigns.
To overcome this, our algorithm employs an advanced multi-model consensus voting framework. This innovative approach aggregates the binding poses and scores predicted by multiple independent docking models. It then selectively outputs only those molecular structures that achieve a high consensus—both a consistently predicted binding mode and a high binding score across the majority of models.
This rigorous consensus mechanism acts as a powerful filter, dramatically reducing the false positive rate and substantially increasing the success rate of lead identification. Validation studies demonstrate that the CrossDock module achieves a 3.5-fold higher hit rate and enrichment factor compared to leading commercial and open-source docking software, establishing it as a superior tool for high-throughput virtual screening.